





 pace and military technologies were the forerunners of many image processing applications which later found their way into medical imaging.
pace and military technologies were the forerunners of many image processing applications which later found their way into medical imaging.
One of the most important was the Landsat program of NASA. It created sets of images of the earth consisting of four or more images of different spectral windows (usually, two within the visible spectrum and two within the infrared spectrum).
Similar approaches are used today in medical image processing. Image segmentation is one of the most important tools in automated image analysis.
 Plain and postcontrast T1-weighted, T2-weighted, and diffusion images can be used as multispectral images. Reducing the representation of an image to a small number of components was one of the image-processing projects based on such pictures, a process called feature extraction.
Plain and postcontrast T1-weighted, T2-weighted, and diffusion images can be used as multispectral images. Reducing the representation of an image to a small number of components was one of the image-processing projects based on such pictures, a process called feature extraction.
It permits the separation of the basic parts of an image by sets of features that can be extracted from the image (or several images) and, in turn, then can be used to calculate other features such as edges and textures.
Segmentation is also applied in preprocessing of images for multimodality image registration. Image segmentation can be used in static images and, quite important for the use of contrast agents, in dynamic time-varying images.
The detection of gray-level discontinuity allows the highlighting of points, lines, and edges in an image [⇒ Bezdek 1993, ⇒ Lundervold 1992].
Similarity techniques reveal areas of similar signal intensities using thresholding, region growing, as well as region splitting and merging.
An overview of the components of an image-analysis system is given in Figure 15-10. A detailed description of segmentation is beyond the limits of this chapter, but is available in other treatises [⇒ Gonzalez 2008, ⇒ Clarke 1993, ⇒ Russ 2017].
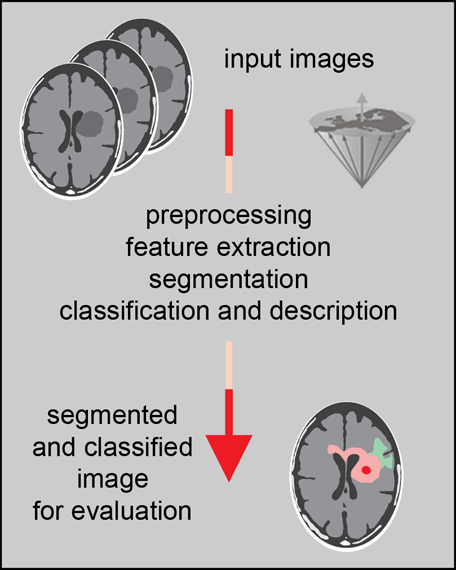
Figure 15-10:
Steps in image analysis: preprocessing improves the quality of the image by reducing artifacts; feature extraction and selection provide the measurement vectors on which segmentation is based. After segmentation, classification and description allow pattern recognition.
Multispectral models can be divided into supervised and unsupervised models. An unsupervised classification (like cluster analysis) into connected regions is generally sufficient to provide good partition of an image into relevant component structures. Supervised pattern recognition is mainly successful where a reliable classification can be expected on the basis of a priori knowledge of the tissue parameters [⇒ Alaux 1990].
The classical example is gray and white matter separation on the basis of relaxation time data (Figure 15-11).
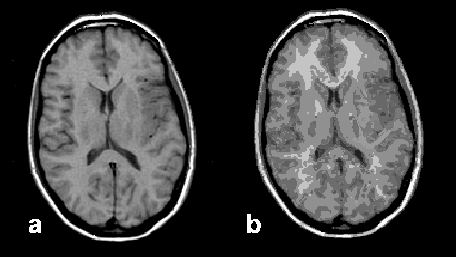
Figure 15-11:
Image segmentation: (a) original brain image, and (b) segmented image presenting 90 different tissue components.
 Practical Applications. In medicine, segmentation is applied for the division of images into components reflecting the same or similar tissues.
Practical Applications. In medicine, segmentation is applied for the division of images into components reflecting the same or similar tissues.
Today, the concept of segmentation and its application of volumetry have become fast and clinically usable. Segmentation allows identification of anatomical areas of interest for diagnosis and therapy, for instance for planning of surgery.
Measurement of tumor volume before and after treatment has become a relatively easy task with image segmentation. Among other applications are quantitative measurements of brain atrophy in patients with Alzheimer's disease or alcoholic brain damage.
In cardiac MR imaging segmentation methods and contour-detection methods have been successfully applied to detect the borders of myocardium in order to calculate parameters like ejection fraction and myocardial mass. Automatic contour detection can be used for the three-dimensional depiction of bone or soft tissue structures, e.g., to produce prostheses.